
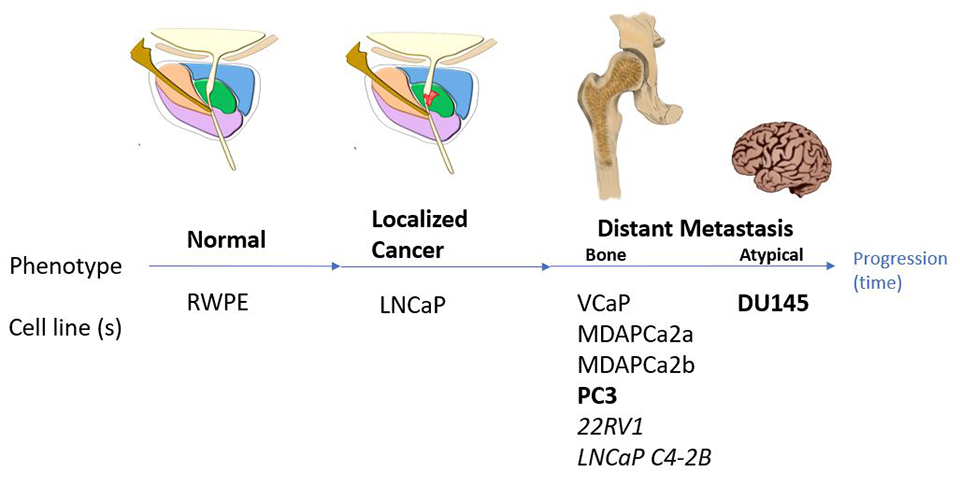
In our recent study (San Martin et.al. 2021), we have characterized the 3D genome structure of a cohort of prostate cancer cell lines that model the continuum of prostate cancer, from normal epithelium to metastatic.
We have identified a cohort of about 400 genes that change chromatin compartmentalization, from open chromatin (A compartment) to close heterochromatin (B compartment), characterizing 60 gene clusters that change not only compartments but also topological associating domains (TADs) distribution and mRNA expression.
We have also determined that significant changes in compartment identity is concurrent with changes in 3D genome architecture at the local level: from distinct TAD structure to no structure and vice versa, shift to structural deserts, or increase of stalled loops.
- GEO link to data in this study
- Genome Browser tracks related to this study
- HiGlass visualizing tool for Hi-C datasets.
- Supplementary Data Files