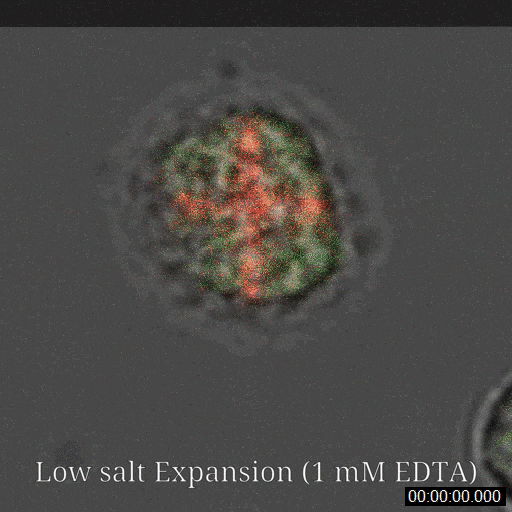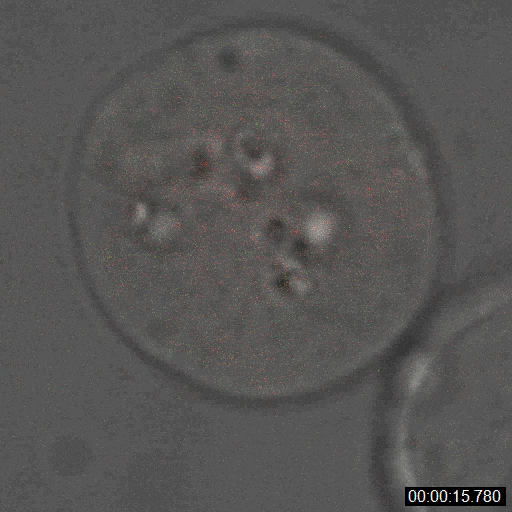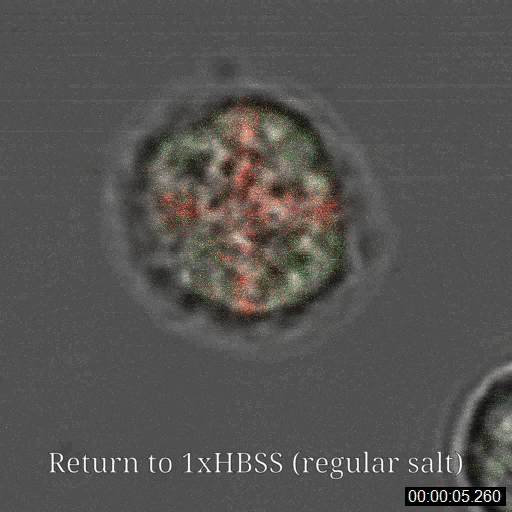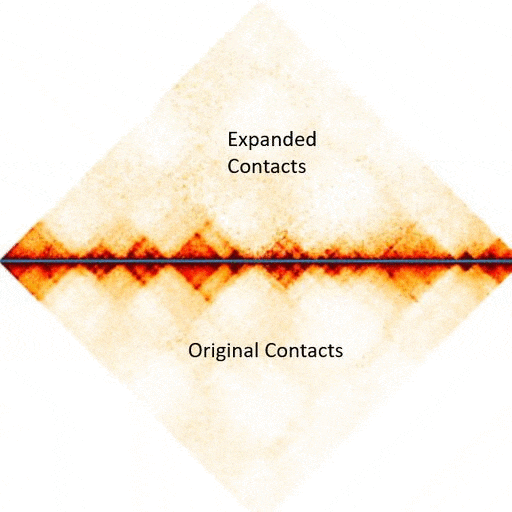https://www.ncbi.nlm.nih.gov/myncbi/rachel.mccord.1/bibliography/public/
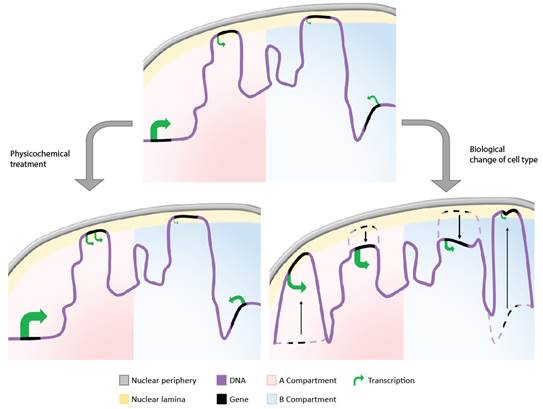
Das P, San Martin R, McCord RP. Differential contributions of nuclear lamina association and genome compartmentalization to gene regulation. Nucleus. (2023).
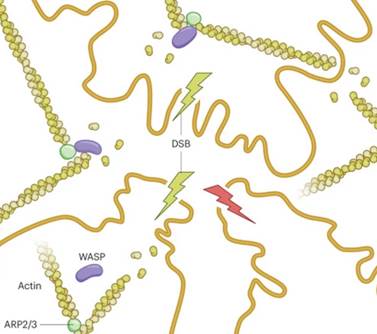
Li H, McCord RP. Actin up: shifting chromosomes toward repair, but also translocations. Nat Struct Mol Biol. (2023).
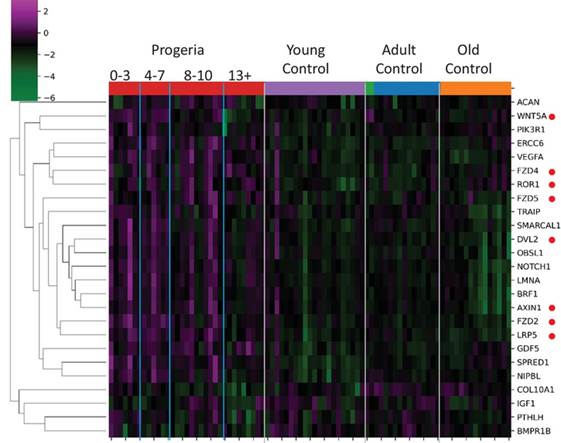
San Martin R, Das P, Sanders JT, Hill AM, McCord RP. Transcriptional profiling of Hutchinson-Gilford Progeria syndrome fibroblasts reveals deficits in mesenchymal stem cell commitment to differentiation related to early events in endochondral ossification. eLife. (2022).
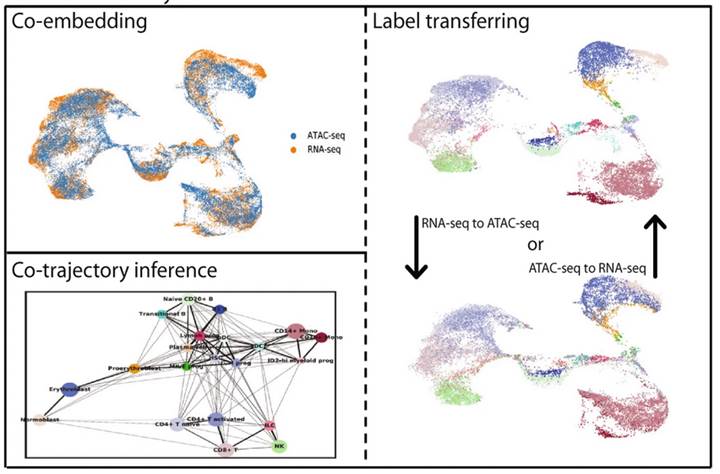
Xu Y, Begoli E, McCord RP. sciCAN: single-cell chromatin accessibility and gene expression data integration via cycle-consistent adversarial network. NPJ Syst Biol Appl. (2022).
Das P, Shen T, McCord RP. Characterizing the variation in chromosome structure ensembles in the context of the nuclear microenvironment. PLoS Comp. Bio. (2022).
Sanders JT, Golloshi R, Das P, Xu Y, Terry PH, Nash DG, Dekker J, McCord RP. Loops, topologically associating domains, compartments, and territories are elastic and robust to dramatic nuclear volume swelling. Scientific Reports. (2022).
Xu Y, McCord RP. CoSTA: unsupervised convolutional neural network learning for spatial transcriptomics analysis. BMC Bioinformatics. (2021).
Xu Y, Das P, McCord RP. SMILE: mutual information learning for integration of single-cell omics data. Bioinformatics. (2022).
San Martin R, Das P, DosReis R, Xu Y, McCord RP. Alterations in chromosome spatial compartmentalization classify prostate cancer progression. J Cell Biol 221 (2): e202104108. (2022).
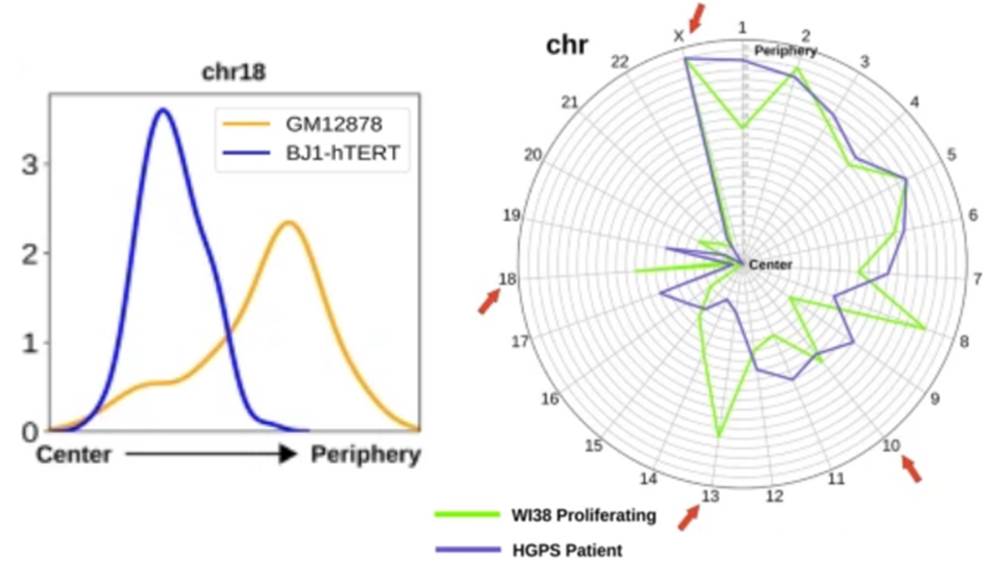
Das P, Shen T, McCord RP. Inferring chromosome radial organization from Hi-C data. BMC Bioinformatics 10;21(1):511. (2020).
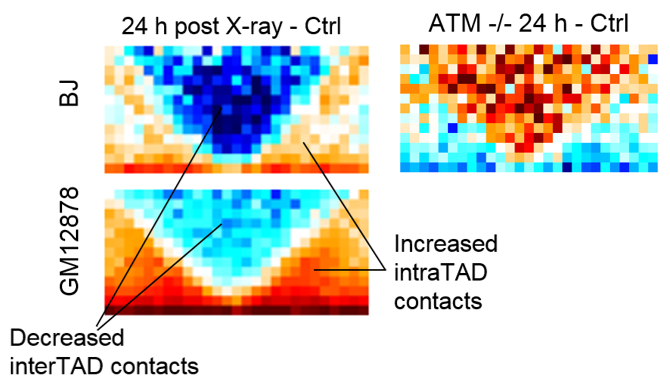
Sanders JT, Freeman TF, Xu Y, Golloshi R, Stallard MA, San Martin R, Balajee AS, McCord RP. Radiation-induced DNA damage and repair effects on 3D genome organization. Nature Communications 11, 6178. (2020).
Golloshi R, Playter C, Freeman TF, Das P, Raines TI, Garretson JH, Thurston D, McCord RP. Constricted migration is associated with stable 3D genome structure differences in cancer cells. EMBO Reports. (2022).
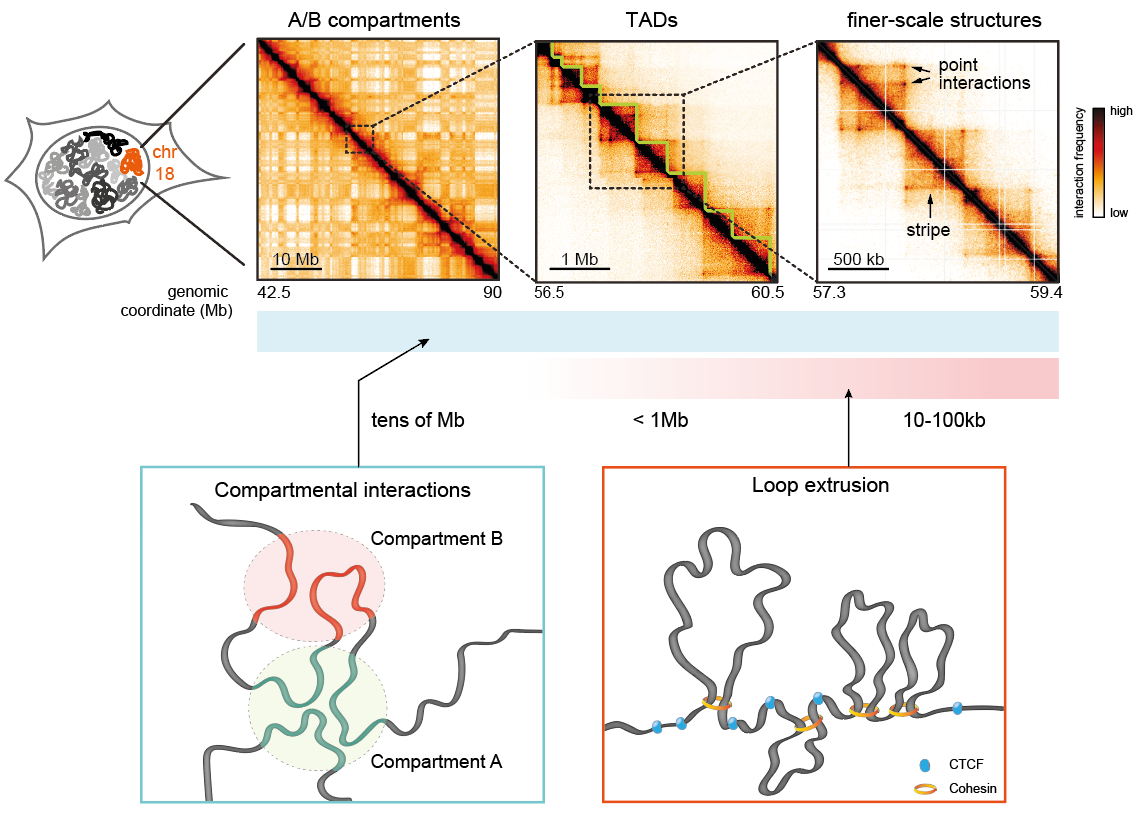
McCord RP, Kaplan N, Giorgetti L. Chromosome Conformation Capture and Beyond: Toward an Integrative View of Chromosome Structure and Function. Molecular Cell. (2020).
Das P, Golloshi R, McCord RP, and Shen T Using contact statistics to characterize structure transformation of biopolymer ensembles. Physical Review E. (2020).
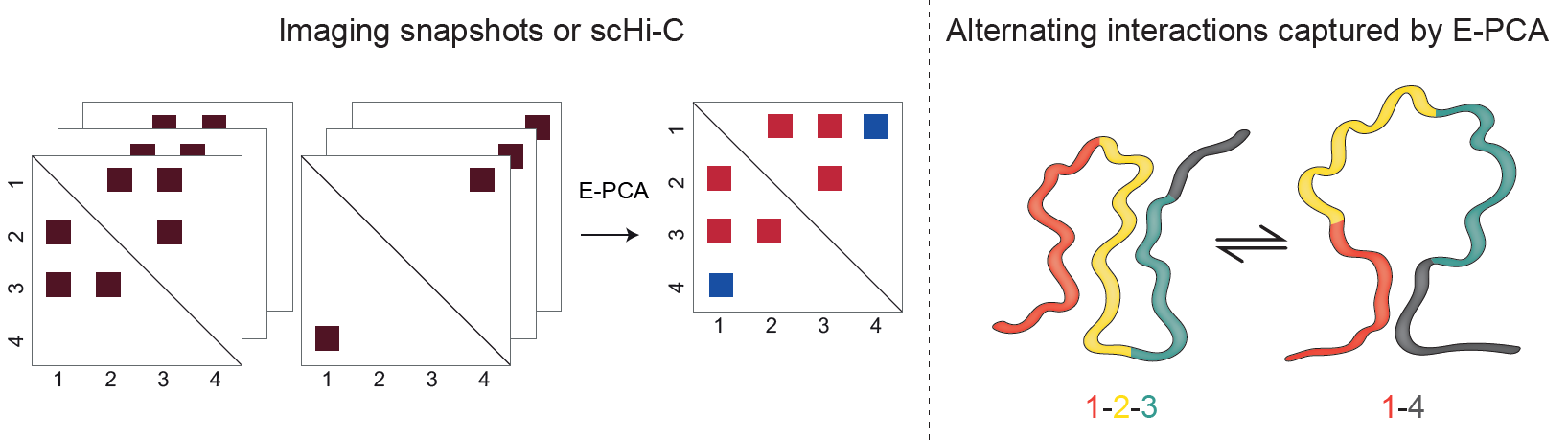
Lindsay RJ, Pham B, Shen T, McCord RP. Characterizing the 3D structure and dynamics of chromosomes and proteins in a common contact matrix framework. Nucleic Acids Research. (2018).
Golloshi R, Sanders JT, McCord RP. Iteratively improving Hi-C experiments one step at a time. Methods. (2018).
McCord RP. Chromosome biology: How to build a cohesive genome in 3D. Nature 551, 38–40. (2017).
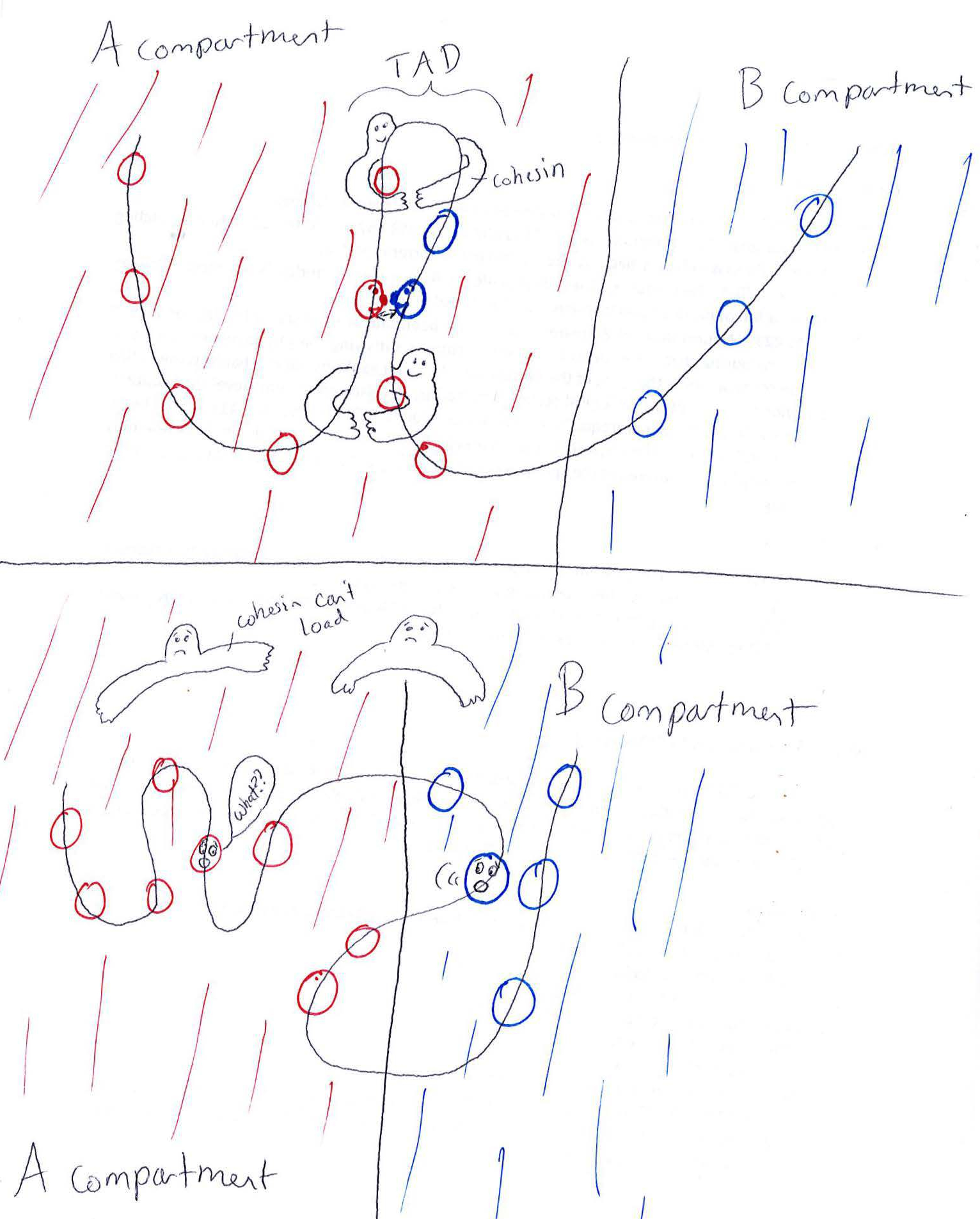
This is the original drawing I sent Nature to explain my figure idea. You can see the professional-looking version they produced at the article link– without the fun faces!
Golloshi R, Sanders JT, McCord RP. Genome organization during the cell cycle: unity in division. Wiley Interdiscip Rev Syst Biol Med. (2017)
McCord RP, Nazario-Toole A, Zhang H, Chines PS, Zhan Y, Erdos MR, Collins FS, Dekker J, Cao K. Correlated alterations in genome organization, histone methylation, and DNA-lamin A/C interactions in Hutchinson-Gilford progeria syndrome. Genome Res. 23(2):260-9 (2013)
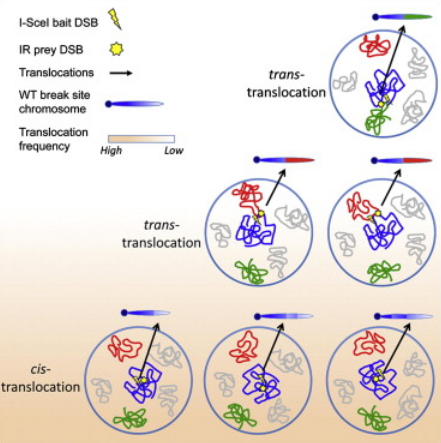
Zhang Y*, McCord RP* , Ho YJ, Lajoie BR, Hildebrand DG, Simon AC, Becker MS, Alt FW, Dekker J. Spatial organization of the mouse genome and its role in recurrent chromosomal translocations. Cell 148(5):908-21.(2012)